The type of mutations tracked matters for maximum sensitivity and specificity
The type of mutations, or genetic variants, tracked in circulating tumor DNA (ctDNA)–based minimal residual disease (MRD) testing matters. Haystack MRD™ prioritizes truncal somatic mutations—those mutations that are present at the initial formation of a tumor, or the trunk of the cancer evolutionary tree. This blog post discusses why preferentially including truncal mutations in MRD test panels is critical for maximizing sensitivity.
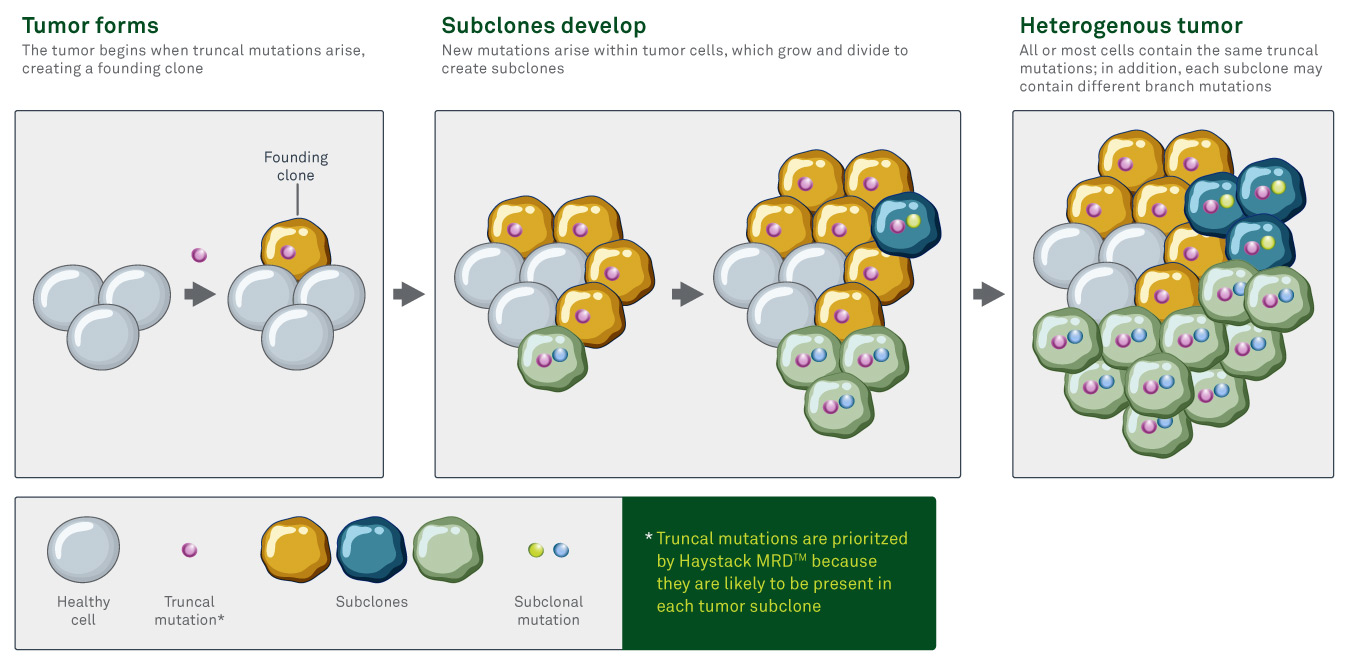
Tumors are heterogenous
Individual cells or populations of cells within a tumor display unique genetic alterations. This spatial heterogeneity is due to the genetic instability of cancer cells as well as differences in the microenvironment (nutrient availability, oxygen, and so on) within the tumor.1
New genetic mutations that occur within an individual cell of the tumor are carried through to the progeny of that cell as the cell divides, creating subclones, or clusters of cells that each contain the new mutation(s). This process occurs throughout a tumor, potentially creating many unique subclones. Each subclone can mutate further over time, creating many genetic branches within the tumor.2
The overall genetic makeup of a tumor can also shift over time. Eventually, one subclone may proliferate faster than others, dramatically shifting the overall genetic makeup of the tumor.2 This temporal heterogeneity can make it difficult to identify tumor cells that have drifted a great degree from the original tumor.
Likewise, metastases may vary substantially from the rest of the tumor.
This heterogeneity presents a theoretical challenge for MRD testing. The gold standard for MRD testing is a tumor-informed approach, where a sample of the patient’s original tumor is sequenced and used for subsequent MRD testing. Because of the heterogenous nature of the tumor, the sample that is used to design the test may not represent the genetic makeup of the entire tumor and any residual disease that is left behind. If that residual disease stems from a subclone that is substantially different than the original sample, the residual cells may be more difficult to detect if truncal mutations are not prioritized in the test design.
Truncal mutations are intrinsically resistant to tumor heterogeneity
Truncal mutations are genetic alterations that occur early in the development of a tumor and are theoretically carried through to all cells within the tumor. These mutations are often early drivers of oncogenesis, but they can also include passenger mutations that arise by chance and do not influence tumor formation.
Truncal mutations often arise in genes that regulate processes critical to tumor development. For example, in colon cancer, the following genes are frequently mutated to drive the initial formation of the tumor:
- TP35 encodes transcription factor p53, which regulates genetic stability, cell proliferation, cell survival, invasion, and more3
- The RAS gene family (especially KRAS) encodes RAS proteins, which mediate cell survival4
- The APC gene product affects cell proliferation, cell survival, and cell adhesion, among other functions5
In contrast, branch or subclonal mutations are those that arise later in tumor development and are only present in a subset of tumor cells.
Truncal mutations are intrinsically resistant to tumor heterogeneity because they tend to be carried through to all subclones within the tumor. That’s why truncal mutations make the best markers for MRD. If residual or recurrent disease comes from a highly divergent subclone, it is more likely to be identified if the test panel focuses on truncal mutations rather than on branch mutations that may not exist in that specific subclone.
Demonstrated clinical value of testing for truncal mutations
Several studies have shown that prioritizing truncal mutations enhances the sensitivity of ctDNA-based MRD testing.
- A study in breast cancer patients showed that truncal mutations have higher circulating levels than branch mutations and are less likely to disappear from tumors during follow-up testing6
- A study that optimized the detection of ctDNA found that branch mutations were more likely than truncal mutations to disappear over time: the longer the time between blood draws, the less likely a branch mutation observed at an early time point would also be detected at a later time point, whereas truncal mutations were more stable over time7
- In another study, truncal mutations were detected in liquid biopsies more easily than other types of mutations, presumably due to their broad prevalence (high variant allele frequency) within the tumor8
Haystack MRD prioritizes truncal mutations
When developing each patient’s personalized testing panel, Haystack MRD’s mutation-selection algorithm preferentially includes truncal mutations. By prioritizing mutations that are known to drive the patient’s disease and are present in both the primary tumor as well as any distant metastases, Haystack MRD can provide a more accurate representation of overall disease burden, resulting in more confident MRD results. The prioritization of truncal mutations contributes to Haystack MRD’s best-in-class sensitivity with the ability to detect 1 mutant molecule out of 1 million molecules of normal DNA.
References
- Gerlinger, M, Rowan, AJ, Horswell, S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. New Engl J Med, 2012;366(10):883-892. doi:10.1056/nejmoa1113205
- Liebl MC, Hofmann TG. The role of p53 signaling in colorectal cancer. Cancers (Basel). 2021;13(9):2125. doi:10.3390/cancers13092125
- Saeed O, Lopez-Beltran A, Fisher KW, et al. RAS genes in colorectal carcinoma: pathogenesis, testing guidelines and treatment implications. J Clin Pathol. 2019;72(2):135-139. doi:10.1136/jclinpath-2018-205471
- Zhang L, Shay JW. Multiple roles of APC and its therapeutic implications in colorectal cancer. J Natl Cancer Inst. 2017;109(8). doi:10.1093/jnci/djw332
- Murtaza M, Dawson SJ, Pogrebniak K, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun. 2015;6(1). doi:10.1038/ncomms9760
- Li S, Noor ZS, Zeng W, et al. Sensitive detection of tumor mutations from blood and its application to immunotherapy prognosis. Nat Commun. 2021;12(1):1-14. doi:10.1038/s41467-021-24457-2
- Koba H, Kimura H, Yoneda T, et al. Molecular features of tumor-derived genetic alterations in circulating cell-free DNA in virtue of autopsy analysis. Sci Rep. 2021;11(1):8398. doi:10.1038/s41598-021-87094-1