Haystack’s personalized, tumor-informed MRD testing yields superior results
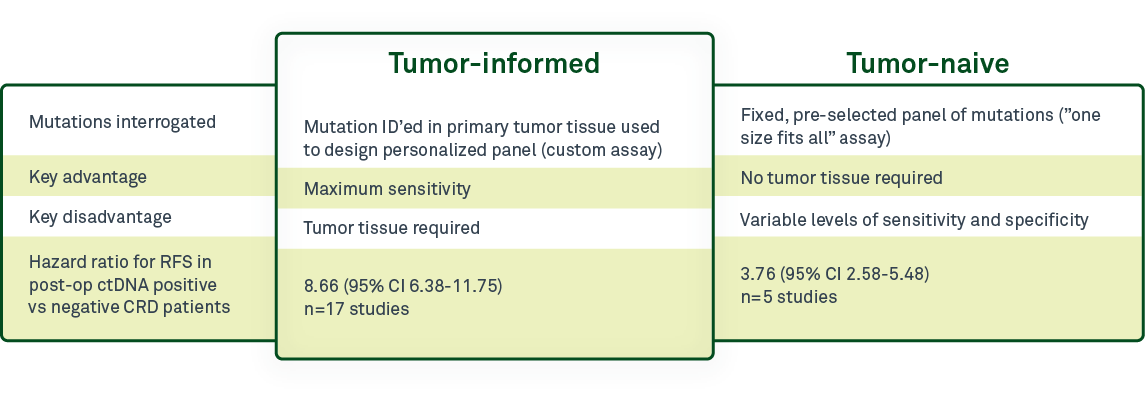
One of the most critical differentiators among minimal residual disease (MRD) tests is whether they are tumor-informed—that is, using information from a patient’s tumor sample to design a personalized test—or whether they are tumor-naive, using the same, “one size fits all” assay for all patients. It is important to understand the difference between these 2 test designs for circulating tumor DNA (ctDNA)-based MRD testing and the advantages and disadvantages of each approach.
Tumor-naive testing
In tumor-naive, or tumor-agnostic, MRD testing, the same preselected panel of mutations is analyzed for all patients. The advantage is that this type of testing requires no tumor tissue. The tradeoff is the lack of personalization with this testing approach. Because the test panel is the same for all patients, only a subset of the mutations in the test panel are actually present in each patient’s residual or recurrent disease. As a result, sensitivity will be lower than with tumor-informed tests.
Tumor-naive approaches can be used to monitor recurrence in patients for whom an original tumor sample was never collected. If ctDNA monitoring is initiated years after the original tumor is removed, and no tumor sample is available, a tumor-naive test would be the only option available to that patient. However, the early detection that is required for MRD testing requires exquisite sensitivity—therefore, tumor-naive testing is not as suitable as tumor-informed testing for identifying residual, recurrent, or resistant disease.
Tumor-informed testing
In contrast, tumor-informed assays, such as Haystack MRD™, use a tumor tissue sample to design a personalized assay for each patient based on the mutations present in that patient’s tumor. The sample tissue can be obtained during surgical resection, or in some cases, from a biopsy.
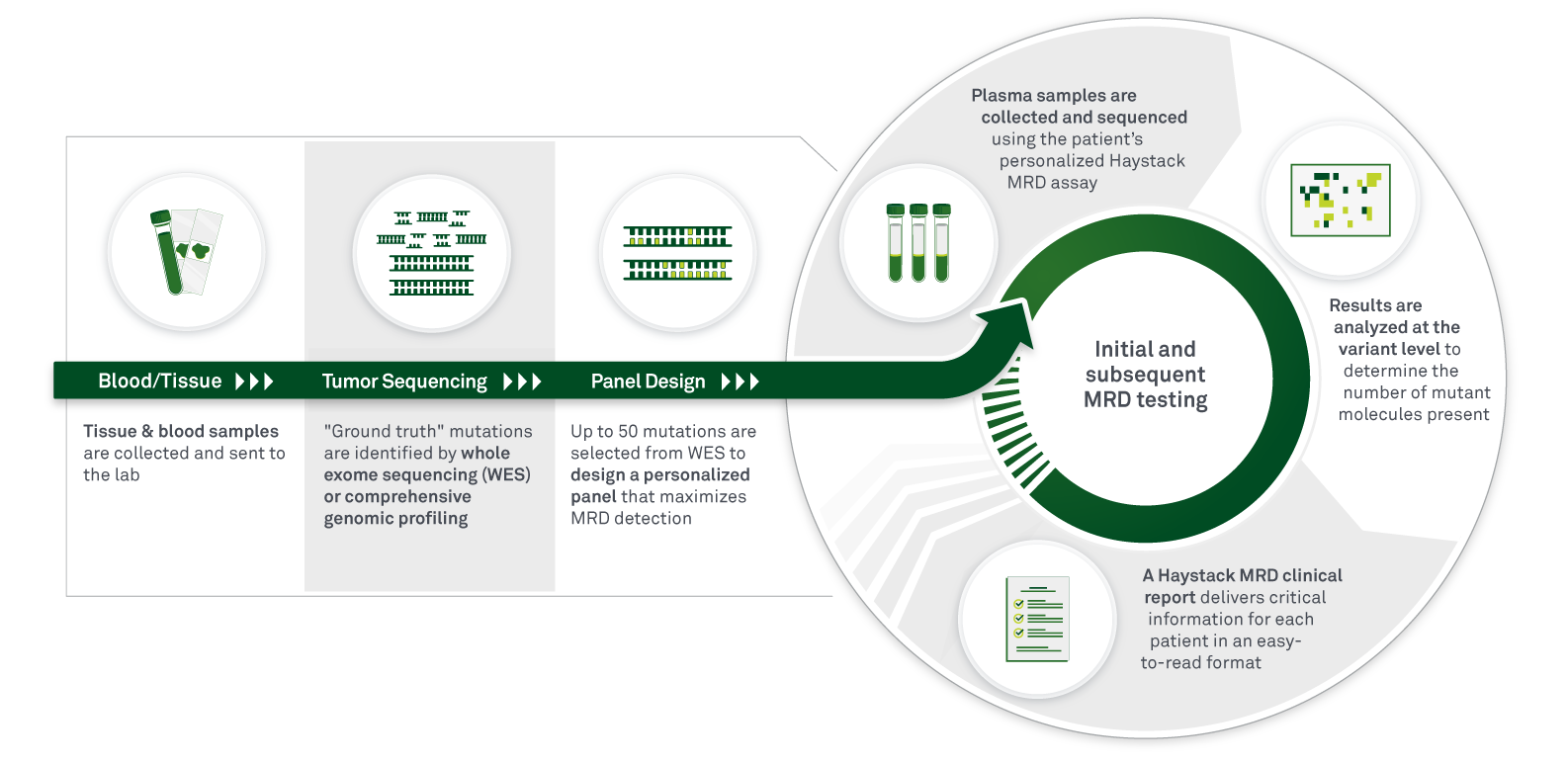
While tumor-informed testing may require a longer turnaround time, the key advantage to this method is that maximum sensitivity can be achieved. Because the mutations being tracked are patient-specific, mutations are now included that may not have been part of a tumor-naive panel, and non-tumor CHIP (clonal hematopoiesis of indeterminate potential) mutations can be filtered out, minimizing the chances of false-positive or false-negative results.
The resulting ultrahigh sensitivity allows for detection of the lowest levels of ctDNA, and therefore, detection at the earliest timepoints, meaning that treatment can begin as soon as possible when tumor burden is still low. Tumor-informed MRD may be especially beneficial for heterogenous tumors, where a generic test is less likely to capture the range of possible mutations in a patient’s tumor.
In addition, the longer turnaround time for tumor-informed tests is only a factor for the initial test design. Once the personalized assay is created, that test is used for all subsequent follow-up time points, placing the turnaround time for subsequent testing on par with tumor-naive testing, while having the advantage of being more sensitive. In other words, after the test panel is designed, there is little benefit to using a tumor-naive approach.
Clinical studies demonstrate increased sensitivity of tumor-informed approach
A 2022 meta-analysis by Chidharla et al1 that evaluated 23 studies including 3568 patients with stage I-IV colorectal cancer demonstrated improved utility of tumor-informed testing over tumor-naive. Tumor-informed tests were more effective than tumor-naive at predicting recurrence-free survival following a positive ctDNA result, with a hazard ratio (HR) of 8.66 for tumor-informed methods and 3.76 for tumor-naive testing.
Other studies have shown that tumor-informed approaches were more sensitive than tumor-naive ones for pancreatic cancer2 and breast cancer3 patients, particularly for detection of low levels of ctDNA. The detection rate for ctDNA in stage 0-IV pancreatic cancer patients after surgical resection and without neoadjuvant therapy was 39% for tumor-naive and 56% for tumor-informed approaches.2
Market trends
Analysts predict that the MRD market will trend toward tumor-informed approaches in the next few years. They indicate that the potential logistical barriers to tumor-informed testing have not deterred oncologists from ordering these tests.4 They also predict that by 2027, most oncologists will choose tumor-informed approaches.5
Summary
The choice between tumor-informed and tumor-naive ctDNA testing depends on the clinical context, testing goals, and tumor tissue availability.
While tumor-naive testing has the benefit of faster turnaround for the initial test because of the lack of requirement for tumor tissue, tumor-informed approaches, such as Haystack MRD, offer maximum sensitivity and specificity and demonstrated clinical benefit over tumor-naive testing.
To offer the greatest benefit of MRD detection, tests need to be able to identify the lowest levels of ctDNA. Tumor-informed Haystack MRD provides the highest sensitivity and specificity to allow accurate monitoring of treatment response and disease clearance and to detect the earliest stages of disease relapse, thereby delivering insights at critical moments for important treatment decisions.
References
- Chidharla A, Rapoport E, Agarwal K, et al. Circulating tumor DNA as A MRD assessment and recurrence risk in patients undergoing curative intent resection with or without adjuvant chemotherapy in colorectal cancer: A meta-analysis. bioRxiv. Published online 2022. doi:10.1101/2022.11.04.22281967
- Watanabe K, Nakamura T, Kimura Y, et al. Tumor-informed approach improved ctDNA detection rate in resected pancreatic cancer. Int J Mol Sci. 2022;23(19):11521. doi:10.3390/ijms231911521
- Santonja A, Cooper WN, Eldridge MD, et al. Comparison of tumor-informed and tumor-naive sequencing assays for ctDNA detection in breast cancer. EMBO Mol Med. 2023;15(6):e16505. doi:10.15252/emmm.202216505
- [DeciBio Consulting. Oncology Liquid Biopsy (LBx) Market Report 2022–2027]
- [Westenberg, D and Petersen A. Piper Sandler. MRD Total Addressable Market, October 26, 2022]


